Z-DNA is one of the many possible structures of the DNA double helix, is a left-twisted double helix (as opposed to right-twisted, as the most common form of ). Z-DNA is one of the three biologically active double helix structures of DNA, along with A-DNA and B-DNA, although its exact functions have not been determined to date [1] .
Study History
Left-handed DNA was first discovered by Robert Wells and colleagues when studying a polymer formed by repetitions of inosine - cytosine [2] . They observed a “reverse” circular dichroism in such DNA, from which they made the correct conclusion that its chains twist each other in the direction to the left. Subsequently, the crystal structure of Z-DNA was published, where X-ray diffraction analysis revealed that it is the first single-crystal DNA fragment ( self- complementary DNA hexamer d (CG) 3 ). It was found that Z-DNA is a left-handed double helix of DNA from two anti-parallel chains connected by bonds between pairs of nitrogenous bases . These works were carried out by Andrew Wang ( eng. Andrew Wang ), Alexander Rich and their staff at the Massachusetts Institute of Technology [3] .
In 1970, it was shown that the most common B-form DNA can become Z-form. In this experiment, it was demonstrated that the circular dichroism of the polymer (dG-dC) in ultraviolet rays with a solution of 4 M NaCl was changed to the exact opposite [4] . The fact that during this transition the B-form passed into the Z-form was confirmed by the results of Raman spectroscopy [5] . The crystallization of compound B- and Z-DNA, carried out in 2005 [6] , gave a better understanding of the potential role that Z-DNA plays in the cell . Wherever there are segments of Z-DNA forms, there should also be B-Z-compounds at their ends connecting the Z-form with the B-form found throughout the rest of the genome .
In 2007, RNA- version of Z-DNA was described as a transformed form of the double right - twisted A-RNA helix into a left-handed helix [7] . The transition from A-RNA to Z-RNA , however, was already described in 1984 [8] .
Structure
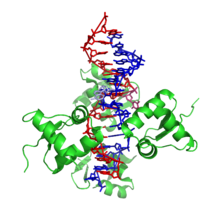
Z-DNA is significantly different from right-handed forms. Z-DNA is left-handed and has a primary structure , repeated every 2 base pairs. There are 12 base pairs per turn of the helix. In contrast to A- and B-DNA, in Z-DNA the major groove is poorly distinguishable, the minor groove is narrow and deep [9] . In general, the structure of Z-DNA is energetically unfavorable, although some conditions may activate its formation, such as: alternating purine - pyrimidine sequences (especially poly (dGC) 2 ), negative supercoiling of DNA , high salt content and some cations (all at physiological temperature - 37 ° C and a pH of 7.3-7.4). Z-DNA can bind to B-DNA into a structure that leads to the displacement of base pairs (see fig.) [10] .
Another feature of Z-DNA is the alternation of conformations of nucleotide residues. Deoxycytidine is in a standard conformation: sugar in C2'-endoconformation (see fig.), And the base is in anti- conformation (that is, the base is turned in the direction opposite to the hydroxyl group at the fifth carbon atom; in this position are the bases in the polynucleotide chain [11] ). In deoxyguanosine, sugar is in C3'-endoconformation , and the base has an extremely atypical syn- conformation [12] .
Base stacking in Z-DNA has new, intrinsic properties only. Thus, stacking interactions exist only between the cytosine residues of opposite chains, and guanine residues do not interact at all with each other [1] .
Phosphates in Z-DNA are not equivalent to each other and are removed at different distances from the axis of the helix; for guanine nucleotides, this distance is 0.62 nm , and for cytosine nucleotides - 0.76 nm. At the same time, the neighboring sugars “look” in opposite directions, and because of this, the line that sequentially connects the phosphorus atoms in the chain becomes zigzag (hence the name Z-DNA) [1] .
The structure of Z-DNA is difficult to study, because it practically does not exist in the stable form of a double helix. In contrast, the left-handed Z-DNA helix is a temporary structure that appears as a result of biological activity and quickly disappears [13] .
Transition from B-DNA to Z-DNA
As already mentioned, B-and Z-forms are able to go into each other. This occurs when the ionic strength of the solution or the concentration of cations that neutralize the negative charge of the phosphodiester scaffold changes. At the same time, there is no need for the transition to diverge the chains, it is initiated by the breaking of hydrogen bonds at several base pairs, after which the guanine is fixed in the synconformation, the hydrogen bonds are restored, and the bases again form Watson-Crick pairs . The transition region moves in a spiral in the form of a loop [1] .
Predicting Z-DNA Structure
Currently, it is possible to predict a plausible DNA sequence in the form of Z-DNA. An algorithm for predicting the propensity of DNA to rearrange from the B-form to the Z-form, ZHunt , was written in 1984 by Dr. P. Shing Ho of the Massachusetts Institute of Technology [14] . Later, this algorithm was developed by Tracy Camp and colleagues to determine the formation of Z-DNA in the entire genome [15] .
The ZHunt algorithm is available at Z-Hunt online .
Biological significance
Z-DNA was found in representatives of all three domains of life: archaea (in particular, in haloarheans [16] ), bacteria and eukaryotes [9] . So far, no clear biological functions of Z-DNA have been identified, however, it is believed to be involved in the regulation of gene expression at the transcription level. Indeed, it is reliably known that the regulation of gene expression in eukaryotes is associated with the sequence dm 5 -dG, which is in physiological conditions in the form of Z-DNA. This regulation may be mediated by supercoiling , binding to proteins specific for Z-DNA, determined by type cations and deoxycytidine methylation [17] .
The assumption that Z-DNA provides for DNA supercoiling during transcription [6] [18] is supported by the fact that the potential for the formation of Z-forms is found in the sites involved in active transcription. A link was shown between the sites of Z-DNA formation in the genes of the 22nd human chromosome and the known sites for the onset of transcription [15] .
Z-DNA is formed after the start of transcription. The first domain , which binds to Z-DNA and has a high affinity for it, was discovered in the enzyme (RNA-specific adenosine deaminase) [19] [20] (this domain is called the Z-alpha domain ). Crystallographic and nuclear magnetic resonance studies have confirmed that this domain binds Z-DNA, regardless of its nucleotide sequence [21] [22] [23] . Similar regions were found in some other proteins homologous to ADAR1 [20] . Identification of the Z-alpha domain formed the basis for characterization of Z-RNA and B-compound with Z-DNA. Studies have shown that the ADAR1 domain that binds Z-DNA allows this enzyme to be localized at the sites of active transcription, where it performs its function - changes the sequence of newly formed RNA [24] [25] .
In 2003, the biophysicist Alexander Rich of the Massachusetts Institute of Technology noticed that the virulence factor of a poxvirus , called E3L, has a Z-alpha-related site that is similar to mammalian protein that binds Z-DNA [26] [27] . In 2005, Rich and colleagues figured out what the value of E3L is for poxvirus. When the expression of E3L genes causes an increase in transcription of several genes of the host cell from 5 to 10 times, these genes block the ability of cells to self-destruct ( apoptosis ) as a protective reaction against infection .
Rich suggested that Z-DNA is required for transcription and E3L stabilizes Z-DNA, thus increasing the expression of antiapoptotic genes. He also put forward the idea that small molecules can bind to E3L, preventing the protein from combining with Z-DNA, and ultimately interfere with the expression of anti-apoptotic genes. This can potentially be used as a basis for the protection against smallpox caused by pox viruses.
Using antibodies to Z-DNA, this form of DNA was found in the interdisk regions of the polytene chromosomes . The fact is that nucleosomes are only in B-DNA, and the transition to the Z-form destroys the structure of the nucleosome and, therefore, consists of chromatin nucleosomes. In this regard, it is assumed that the Z-form can play a regulatory role, especially since the B → Z transition is reversible [1] .
It has been established that the toxic effect of ethidium bromide on trypanosomes is associated with the transition of their kinetoplast DNA into the Z-form. This effect is due to the EtBr into DNA, due to which DNA loses its native structure, disintegrates, becomes Z-shaped, and because of this becomes incapable of replication [28] .
Comparing the geometric parameters of some forms of DNA
| Geometric parameter | A-form | B-form | Z-form |
|---|---|---|---|
| Direction | right handed | right handed | left handed |
| Repeat unit | 1 pair of bases (p. O.) | 1 p. | 2 n. |
| Turnover (in degrees) | 32.7 ° | 35.9 ° | 60 ° / 2 |
| Bend | 11 n. | 10.5 p. | 12 p. |
| Location p. relative to the axis | + 19 ° | −1.2 ° | −9 ° |
| Lifting along the axis | 2.3 Å (0.23 nm) | 3.32 Å (0.332 nm) | 3.8 Å (0.38 nm) |
| Incline | 28.2 Å (2.82 nm ) | 33.2 Å (3.32 nm) | 45.6 Å (4.56 nm) |
| Torsion | + 18 ° | + 16 ° | 0 ° |
| Base conformation | anti- | anti- | C: anti-, G: syn |
| Sugar conformation | C3'-endo | C2'-endo | C: C2'-endo, G: C3'-endo |
| Diameter | 23 Å (2.3 nm) | 20 Å (2.0 nm) | 18 Å (1.8 nm) |
| Sources: [29] [30] [31] | |||
Notes
- ↑ 1 2 3 4 5 Konichev, Sevastyanova, 2012 , p. 93.
- ↑ Mitsui et al. Physical and enzymatic studies on poly (IC) -poly d (IC), an unusual double-helical DNA (Eng.) // Nature (London): journal. - 1970. - Vol. 228 , no. 5277 . - P. 1166-1169 . - PMID 4321098 .
- ↑ Wang AHJ, Quigley GJ, Kolpak FJ, Crawford JL, van Boom JH, Van der Marel G., Rich A. Molecular structure of a left-handed double (English) // Nature (London) : journal. - 1979. - Vol. 282 , no. 5740 . - P. 680-686 . - DOI : 10.1038 / 282680a0 . - . - PMID 514347 .
- ↑ Pohl FM, Jovin TM Salt-induced co-operative DNA: equilibrium and kinetic studies with poly (dG-dC) (Eng.) // J. Mol. Biol. : journal. - 1972. - Vol. 67 . - p . 375–396 . - DOI : 10.1016 / 0022-2836 (72) 90457-3 . - PMID 5045303 .
- A Thamann TJ, Lord RC, Wang AHJ, Rich A. High salt form of poly (dG-dC) • poly (dG-dC) is left handed Z-DNA: raman spectra of crystals and solutions (English) // Nucl . Acids Res. : journal. - 1981. - Vol. 9 P. 5443-5457 . - DOI : 10.1093 / nar / 9.20.5443 . - PMID 7301594 .
- 2 1 2 Ha SC, Lowenhaupt K., Rich A., Kim YG, Kim Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases (Eng.) // Nature: journal. - 2005. - Vol. 437 , no. 7062 . - P. 1183-1186 . - DOI : 10.1038 / nature04088 . - . - PMID 16237447 .
- ↑ Placido D., Brown BA 2nd, Lowenhaupt K., Rich A., Athanasiadis A. A left-handed RNA double-handed by RAL-editing enzyme ADAR1 (English) // Structure: journal. - 2007. - Vol. 15 , no. 4 P. 395-404 . - DOI : 10.1016 / j.str.2007.03.001 . - PMID 17437712 .
- ↑ Hall K., Cruz P., Tinoco I Jr, Jovin TM, van de Sande JH 'Z-RNA' - a left-handed RNA double helix (Eng.) // Nature. - 1984. - October ( vol. 311 , no. 5986 ). P. 584-586 . - DOI : 10.1038 / 311584a0 . - . - PMID 6482970 .
- ↑ 1 2 Nelson, Cox, 2008 , p. 281.
- ↑ de Rosa M., de Sanctis D., Rosario AL, Archer M., Rich A., Athanasiadis A., Carrondo MA Crystal structure of a junction between two Z-DNA helices (Eng.) // Proceedings of the National Academy of the United States of America : journal. - 2010. - 18 May ( vol. 107 , no. 20 ). - P. 9088-9092 . - DOI : 10.1073 / pnas.1003182107 . - . - PMID 20439751 .
- ↑ Konichev, Sevastyanova, 2012 , p. 82
- ↑ Konichev, Sevastyanova, 2012 , p. 92
- ↑ Zhang, H., Yu. H., Ren J., Qu. X. Reversible Selective Selective Selective Selective Aggregate (L-aspartic acid complex ) .) // Biophysical Journal : journal. - 2006. - Vol. 90 , no. 9 - P. 3203-3207 . - DOI : 10.1529 / biophysj.105.078402 . - . - PMID 16473901 . Archived October 12, 2008.
- PS Ho PS, Ellison MJ, Quigley GJ, Rich A. A. Z-DNA in naturally occurring sequences (Eng.) // EMBO Journal : journal. - 1986. - Vol. 5 , no. 10 - P. 2737-2744 . - PMID 3780676 .
- ↑ 1 2 Champ PC, Maurice S., Vargason JM, Camp T., Ho PS Distributions of Z-DNA and nuclear factor I in human chromosome 22: a model for coupled transcriptional regulation (Eng.) // Nucleic Acids Res. : journal. - 2004. - Vol. 32 , no. 22 - P. 6501-6510 . - DOI : 10.1093 / nar / gkh988 . - PMID 15598822 .
- ↑ Paul Blum. Archaea: Ancient Microbes, Extreme Environments, and the Origin of Life. - Academic Press, 2001. - Vol. 50. - P. 206. - (Advances in Applied Microbiology).
- ↑ Konichev, Sevastyanova, 2012 , p. 93–94.
- {{{Cite journal - | author = Rich A, Zhang S | year = 2003 - | title = Timeline: Z-DNA: the long road to biological function - | journal = Nature Review | Genetics | volume = 4 | issue = 7 | pages = 566–572 - | pmid = 12838348 | doi = 10.1038 / nrg1115 -}}
- ↑ Herbert A., Rich A. A method to identify and characterize Z-DNA binding proteins using a linear oligodeoxynucleotide (English) // Nucleic Acids Res : journal. - 1993. - Vol. 21 , no. 11 - P. 2669-2672 . - DOI : 10.1093 / nar / 21.11.2669 . - PMID 8332463 .
- 2 1 2 Herbert A., Alfken J., Kim YG, Mian IS, Nishikura K., Rich A. A den adenosine deaminase, double-stranded RNA. (Eng.) // Proceedings of the United States of America : journal. - 1997. - Vol. 94 , no. 16 - P. 8421-8426 . - DOI : 10.1073 / pnas.94.16.8421 . - . - PMID 9237992 .
- ↑ Herbert, A., Schade, M., Lowenhaupt, K., Alfken, J., Schwartz, T., Shlyakhtenko, LS, Lyubchenko, YL, Rich, A. ) // Nucleic Acids Res : journal. - 1998. - Vol. 26 , no. 15 - P. 2669-2672 . - DOI : 10.1093 / nar / 26.15.3486 . - PMID 9671809 .
- ↑ Schwartz, T., Rould, MA, Lowenhaupt, K., Herbert A., Rich A. Z-DNA (English) // Science: journal. - 1999. - Vol. 284 , no. 5421 . - P. 1841-1845 . - DOI : 10.1126 / science.284.5421.1841 . - PMID 10364558 .
- Ade Schade M., Turner CJ, Kühne R., Schmieder P., Lowenhaupt K., Herbert A., Rich A., Oschkinat H. ADAR1 reveals a prepositioned binding surface for Z-DNA (Eng.) // Proceedings of the United States of America : journal. - 1999. - Vol. 96 , no. 22 - P. 2465-2470 . - DOI : 10.1073 / pnas.96.22.12465 . - . - PMID 10535945 .
- Bert Herbert A., Rich A. A. . - 2001. - Vol. 98 , no. 21 . - P. 12132-12113 . - DOI : 10.1073 / pnas.211419898 . - . - PMID 11593027 .
- ↑ Halber D. Scientists' observed activities on left-handed DNA . MIT News Office (September 11, 1999). The date of circulation is September 29, 2008. Archived February 16, 2013.
- ↑ Kim YG, Muralinath M., Brandt T., Pearcy M., Hauns K., Lowenhaupt K., Jacobs BL, Rich A. A role for Z-DNA binding in vaccinia virus pathogenesis (Eng.) // Proceedings of the National Academy of Sciences of the United States of America : journal. - 2003. - Vol. 100 , no. 12 - p . 6974-6979 . - DOI : 10.1073 / pnas.0431131100 . - . - PMID 12777633 .
- ↑ Kim YG, Lowenhaupt K., Oh DB, Kim KK, Rich A. Evidence of vaccinia virulence factor E3L binds to Z-DNA in vivo: Proceedings of the National Academy of Sciences of the United States of America : journal. - 2004. - Vol. 101 , no. 6 - P. 1514-1518 . - DOI : 10.1073 / pnas.0308260100 . - . - PMID 14757814 .
- ↑ Roy Chowdhury, Arnab; Bakshi, Rahul; Wang, Jianyang; Yildirir, Gokben; Liu, Beiyu; Pappas-Brown, Valeria; Tolun, Gökhan; Griffith, Jack D .; Shapiro, Theresa A .; Jensen, Robert E .; Englund, Paul T .; Ullu, Elisabetta. The Killing of African Trypanosomes by Ethidium Bromide (Eng.) // PLoS Pathogens : journal. - 2010. - 16 December ( vol. 6 , no. 12 ). - P. e1001226 . - DOI : 10.1371 / journal.ppat.1001226 .
- ↑ Sinden, Richard R. DNA structure and function. - 1st. - Academic Press, 1994-01-15. - P. 398. - ISBN 0-126-45750-6 .
- ↑ Rich A., Norheim A., Wang AHJ. The chemistry and biology of left-handed Z-DNA (English) // Annual Review of Biochemistry : journal. - 1984. - Vol. 53 , no. 1 . - P. 791-846 . - DOI : 10.1146 / annurev.bi.53.070184.004043 . - PMID 6383204 .
- PS Non-B-DNA structure of d- PS (CA / TG) doesn’t differ from that of Z-DNA (Eng.) // Proceedings of the United States of America : journal. - 1994. - 27 September ( vol. 91 , no. 20 ). - P. 9549-9553 . - DOI : 10.1073 / pnas.91.20.9549 . - . - PMID 7937803 .
Literature
- Konichev A.S., Sevastyanova G. A. Molecular biology. - Publishing center "Academy", 2012. - 400 p. - ISBN 978-5-7695-9147-1 .
- David L. Nelson, Michael M. Cox. Lehninger Principles of biochemistry. - Fifth edition. - New York: WH Freeman and company, 2008. - 1158 p. - ISBN 978-0-7167-7108-1 .