
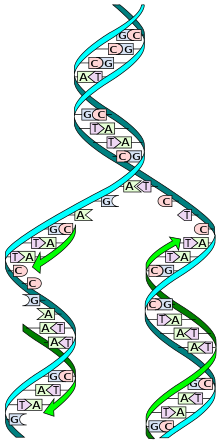
Replication (from lat. Replicatio - renewal) - the process of synthesis of a daughter molecule of deoxyribonucleic acid on the matrix of the parent DNA molecule. During the subsequent division of the mother cell, each daughter cell receives one copy of the DNA molecule, which is identical to the DNA of the original mother cell. This process ensures accurate transmission of genetic information from generation to generation. DNA replication is carried out by a complex enzyme complex, consisting of 15-20 different proteins, called replicoma [1]
Content
Study History
Each DNA molecule consists of one chain of the original parent molecule and one newly synthesized chain. Such a replication mechanism is called semi-conservative. At present, this mechanism is considered proven due to the experiments of Matthew Meselson and Franklin Steel ( 1958 ) [2] . Previously, there were two other models: “conservative” - as a result of replication, one DNA molecule is formed, consisting only of parent chains, and one, consisting only of daughter chains; “Dispersion” - all DNA molecules resulting from replication consist of chains, some parts of which are newly synthesized, while others are taken from the parent DNA molecule. The DNA molecule is cut in half and two patterns are formed. Two templates exit the replication fork. If you imagine them in a straightened form, you can see a ruler of combs that are connected by ends, but have a gap. Imagine that one comb is blue and the other is red. Now we substitute the lower red one (it consists of five ridges, as well as the upper one) with the fifth end to the third upper (third upper needle). Let us extend the chain from above and from below. How would it turn out: five, three, five, etc.- above and below too. Then two more patterns are added to these combs after the patterns (comb) exit the replication fork. From one DNA molecule, two identical maternal (if there are no mutations) molecules are obtained, this is called semi-conservatism.
General
DNA replication is a key event in cell division . It is important that by the time of division of DNA was completely replicated and at the same time only once. This is provided by certain mechanisms of regulation of DNA replication. Replication takes place in three stages:
- replication initiation
- elongation
- replication termination.
Replication regulation is carried out mainly at the initiation stage. This is quite easy to do, because replication can begin not from any part of the DNA, but from a strictly defined site called the replication initiation site . In the genome of such sites there can be just one or many. The concept of replication initiation site is closely related to the concept of replicon . A replicon is a DNA site that contains a replication initiation site and is replicated after the start of DNA synthesis from this site. The genomes of bacteria , as a rule, are one replicon, which means that replication of the entire genome is the result of just one act of replication initiation. Eukaryotic genomes (as well as their individual chromosomes ) consist of a large number of independent replicons, which significantly reduces the total replication time of a single chromosome. Molecular mechanisms that control the number of replication initiation events at each site in a single cell division cycle are called copy control . In addition to chromosomal DNA, bacterial cells often contain plasmids , which are individual replicons. Plasmids have their own copy control mechanisms: they can provide synthesis of just one copy of a plasmid per cell cycle , and thousands of copies [1] .
Replication begins at the site of replication initiation with the unwinding of the DNA double helix, and a replication fork is formed - the site of direct DNA replication. Each site can form one or two replication forks, depending on whether the replication is unidirectional or bi-directional. Bidirectional replication is more common. Some time after the start of replication in an electron microscope, one can observe the replication eye - a region of the chromosome where the DNA is already replicated, surrounded by longer sections of the unreplicated DNA [1] .
In the replication fork, DNA copies a large protein complex (replicom), the key enzyme of which is DNA polymerase . The replication fork moves at a speed of the order of 100,000 pairs of nucleotides per minute in prokaryotes and 500-5000 in eukaryotes [3] .
Molecular Replication Mechanism
Enzymes ( helicase , topoisomerase ) and DNA-binding proteins unwind the DNA, keep the matrix in a diluted state and rotate the DNA molecule. Correct replication is ensured by the exact correspondence of complementary base pairs and the activity of DNA polymerase , which can recognize and correct the error. Prokaryotic replication [ specify ] is carried out by several different DNA polymerases . DNA polymerase I acts on a delayed chain to remove RNA primers and replicate the purified DNA sites. DNA polymerase III is the main DNA replication enzyme that synthesizes the leading strand of DNA and Okazaki fragments in the synthesis of the retarded chain. Then, the synthesized molecules are twisted according to the principle of supercoiling and further DNA compaction. The synthesis of energy-consuming.
The chains of the DNA molecule diverge, form a replication fork , and each of them becomes a matrix on which a new complementary chain is synthesized. As a result, two new double-stranded DNA molecules are formed that are identical to the parent molecule.
- Replication process characteristics
- matrix - the sequence of the synthesized DNA chain is uniquely determined by the sequence of the mother chain in accordance with the principle of complementarity ;
- semi - conservative - one chain of the DNA molecule formed as a result of replication is newly synthesized, and the second is maternal;
- goes from the 5'-end of a new molecule to the 3'-end;
- semi - continuous - one of the DNA chains is synthesized continuously, and the second - in the form of a set of separate short fragments ( Okazaki fragments );
- begins with certain sections of DNA called replication initiation sites ( English origin ) [4] .
Notes
- ↑ 1 2 3 Benjamin Lewin. Chapter 13: The replicon // Genes VIII. - Upper Saddle River, NJ: Pearson Prentice Hall, 2004 .-- ISBN 0131439812 .
- ↑ Matthew Meselson and Franklin W. Stahl. The replication of DNA in Escherichia coli // Proceedings of the National Academy of Sciences of the United States of America : journal. - 1958. - Vol. 44 . - P. 671-682 . - DOI : 10.1073 / pnas . 44.7.671 . - PMID 16590258 .
- ↑ Arthur Kornberg, Tania A. Baker. Chapter 15: The replication mechanisms and operations // DNA replication. - Sausalito, Calif .: University Science Books, 2005 .-- ISBN 1891389440 .
- ↑ N. N. Mushkambarov, S. L. Kuznetsov. Molecular biology. - Medical News Agency, 2007. - ISBN 5-89481-618-1 .